Candida dubliniensis CD36 Genome Snapshot/Overview |
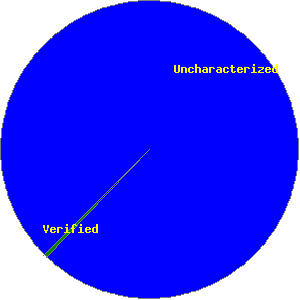
 5834 ORFs, 99.52%
5834 ORFs, 99.52%  28 ORFs, 0.48%
28 ORFs, 0.48% | Feature Type | Total | Chromosome | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Chr1 | Chr2 | Chr3 | Chr4 | Chr5 | Chr6 | Chr7 | ChrR | Nuclear genome | ||
| Total ORFs | 5862 | 1299 | 973 | 727 | 629 | 495 | 417 | 388 | 934 | 5862 |
| Verified ORFs | 28 | 8 | 1 | 6 | 3 | 1 | 2 | 2 | 5 | 28 |
| Uncharacterized ORFs | 5834 | 1291 | 972 | 721 | 626 | 494 | 415 | 386 | 929 | 5834 |
| tRNA | 130 | 23 | 25 | 24 | 17 | 6 | 4 | 1 | 30 | 130 |
| Pseudogenes | 73 | 17 | 7 | 9 | 8 | 8 | 5 | 8 | 11 | 73 |
| Repeat_region | 10 | 1 | 2 | 1 | 2 | 1 | 1 | 2 | 0 | 10 |
| ncRNA | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 2 |
| Total | 6077 | 1340 | 1007 | 761 | 656 | 510 | 427 | 399 | 977 | 6077 |
| Chromosome length (bp) | 14,618,422 | 3,214,061 | 2,289,089 | 1,863,824 | 1,641,709 | 1,245,899 | 1,073,895 | 1,022,435 | 2,267,510 | 14,618,422 |
| Chromosome History | Sequence Updates | Annotation Updates | ||
|---|---|---|---|---|
Total Number | Last Update | Total Number | Last Update | |
0 | N/A | 3 | 2013-05-14 | |
0 | N/A | 2 | 2013-05-14 | |
0 | N/A | 2 | 2013-05-14 | |
0 | N/A | 2 | 2013-05-14 | |
0 | N/A | 3 | 2013-05-14 | |
0 | N/A | 2 | 2013-05-14 | |
0 | N/A | 3 | 2013-05-14 | |
0 | N/A | 2 | 2013-05-14 | |
| Ontology | Details of Annotations | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total Number of Annotations | Graphical View | |||||||||||||||||
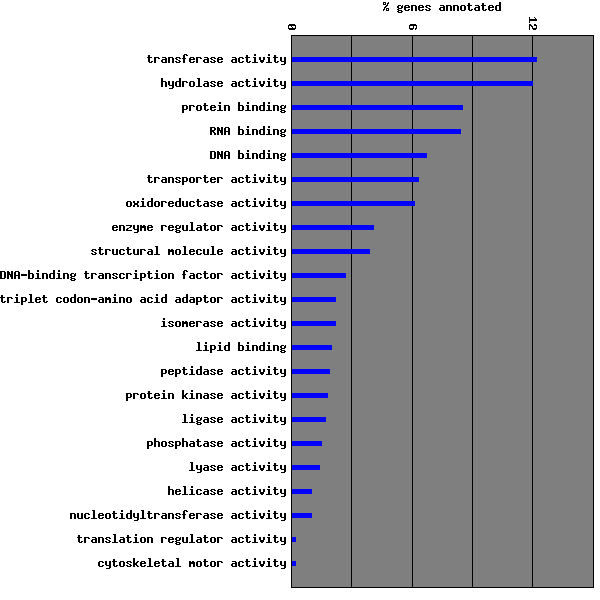
| Molecular Function | 2688 | Go to Molecular Function Graph | ||||||||||||||||
| Cellular Component | 1367 | Go to Cellular Component Graph | ||||||||||||||||
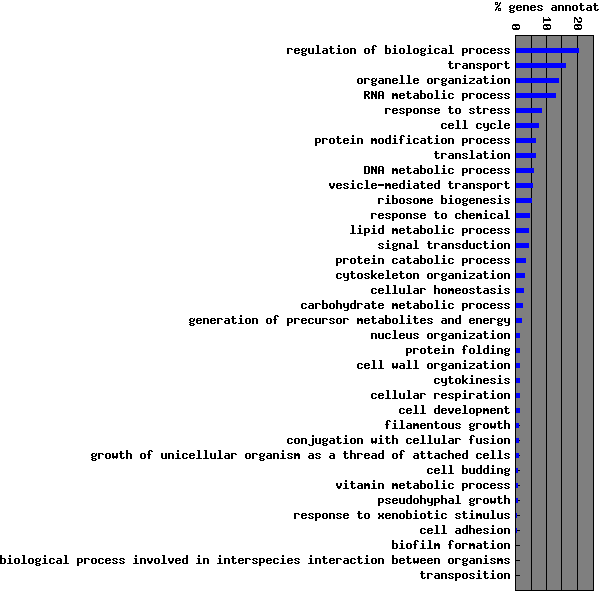
| Biological Process | 2465 | Go to Biological Process Graph | ||||||||||||||||
| All Ontologies | 6520 | |||||||||||||||||

 Return to CGD Return to CGD |
CGD Copyright © 2004-2026 The Board of Trustees, Leland Stanford Junior University. Permission to use the information contained in this database was given by the researchers/institutes who contributed or published the information. Users of the database are solely responsible for compliance with any copyright restrictions, including those applying to the author abstracts. Documents from this server are provided "AS-IS" without any warranty, expressed or implied. To cite CGD, please use the following reference: Skrzypek MS, Binkley J, Binkley G, Miyasato SR, Simison M, Sherlock G (2017). The Candida Genome Database (CGD): incorporation of Assembly 22, systematic identifiers and visualization of high throughput sequencing data. Nucleic Acids Res 45 (D1); D592-D596; see How to cite CGD. |