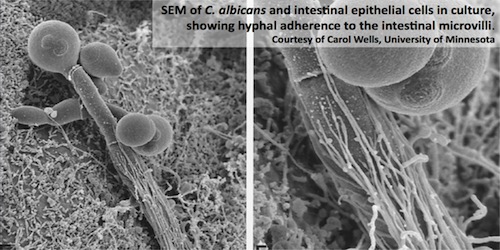
|
|||||||
|
New and Noteworthy |
| |
CGD Survey Needs Your Input |
|---|
This is our chance to improve CGD. Our NIH grant renewal is due in early July and we need your input to make the upgrades that you want to see. If you haven't yet filled out the survey, it takes only 10 to 15 minutes and has great value for all of us. Find it at Candida Genome Database Survey 2024. |
CGD Curation News |
|
CGD Now Has a Newsletter! |
If you didn't receive a copy by email and you'd like one, please send a message to let us know and we'll add you to the member list. Look for the next one in the summer. We will also build an archive for referencing older issues. |
The Fungal Pathogen Genomics Course Now Open for Applications |
The Fungal Pathogen Genomics Course is a hands-on training in web-based data-mining resources for fungal genomes taught by data curation experts. It will be held June 2-7 in Hinxton, UK. The application deadline is Feb 29, 2024 and the course typically fills up. |
The Allied Genetics Conference Now Open for Registration |
TAGC 2024 organized by the Genetics Society of America will be held in Washington, DC on March 6–10, 2024. Abstract submissions are now open until Nov 9 (for presentations) and Jan 4 (for posters only). Registration is now open with early-bird discounts until December 13. |
Expanded Curation of Clinical Data by ITS1 and ITS2 Features for All Species |
Representative copies of ITS1 and ITS2 features for C. auris, C. glabrata, C. dubliniensis, and C. parapsilosis have been added to complement those of C. albicans. The addition of locus pages for these transcribed spacers between rDNA genes (used for strain typing in clinical studies) will allow us to curate these studies by directing the literature to the loci used for identifying the strains. We hope this expansion of searchable clinical data will add value to the database for Candida researchers in medical fields. |
Candida and Candidiasis Conference Now Open for Registration |
Candida and Candidiasis 2023 organized by the Microbiology Society will be held in Montreal, Canada on May 13–17, 2023. Abstract submissions are now open until Feb 17, 2023. Registration is now open with early-bird discounts until April 1. |
New Assembly for Candida glabrata Genome |
We are pleased to announce the incorporation of a new assembly for the C. glabrata CBS138 genome, which leveraged long-read sequencing to correct previous assembly errors in repetitive regions (Xu et al., 2020). The sub-telomeric regions of all chromosomes were substantially lengthened, resulting in changes of chromosomal coordinates for all genomic features. 31 new protein-coding genes were added. The rDNA region on Chr L was expanded significantly, and a second rDNA region was added to Chr M. 62 genes from the previous assembly were removed, and sequences were corrected for an additional 32 features, ranging from single-base changes to major structural rearrangements. The update adds or corrects the sequences of 45 genes encoding GPI-anchored cell wall proteins (GPI-CWPs), which contain many long tandem repeats. |
Archived News |
Click here to view archived news items. |
CGD Copyright © 2004-2024 The Board of Trustees, Leland Stanford Junior University. Permission to use the information contained in this database was given by the researchers/institutes who contributed or published the information. Users of the database are solely responsible for compliance with any copyright restrictions, including those applying to the author abstracts. Documents from this server are provided "AS-IS" without any warranty, expressed or implied. To cite CGD, please use the following reference: Skrzypek MS, Binkley J, Binkley G, Miyasato SR, Simison M, Sherlock G (2017). The Candida Genome Database (CGD): incorporation of Assembly 22, systematic identifiers and visualization of high throughput sequencing data. Nucleic Acids Res 45 (D1); D592-D596; see How to cite CGD. |